One-dimensional steady flow with transport
This problem corresponds to the first problem presented in the MOC3D report Konikow 1996, involving the transport of a dissolved constituent in a steady, one-dimensional flow field. An analytical solution for this problem is given by \cite{wexler1992}. This example is simulated with the GWT Model in \mf, which receives flow information from a separate simulation with the GWF Model in \mf. Results from the GWT Model are compared with the results from the \cite{wexler1992} analytical solution.
Initial setup
Import dependencies, define the example name and workspace, and read settings from environment variables.
[1]:
import os
import pathlib as pl
import flopy
import git
import matplotlib.pyplot as plt
import numpy as np
from flopy.plot.styles import styles
from modflow_devtools.misc import get_env, timed
# Example name and workspace paths. If this example is running
# in the git repository, use the folder structure described in
# the README. Otherwise just use the current working directory.
example_name = "ex-gwt-moc3dp1"
try:
root = pl.Path(git.Repo(".", search_parent_directories=True).working_dir)
except:
root = None
workspace = root / "examples" if root else pl.Path.cwd()
figs_path = root / "figures" if root else pl.Path.cwd()
# Settings from environment variables
write = get_env("WRITE", True)
run = get_env("RUN", True)
plot = get_env("PLOT", True)
plot_show = get_env("PLOT_SHOW", True)
plot_save = get_env("PLOT_SAVE", True)
Define parameters
Define model units, parameters and other settings.
[2]:
# Scenario-specific parameters - make sure there is at least one blank line before next item
parameters = {
"ex-gwt-moc3d-p01a": {
"longitudinal_dispersivity": 0.1,
"retardation_factor": 1.0,
"decay_rate": 0.0,
},
"ex-gwt-moc3d-p01b": {
"longitudinal_dispersivity": 1.0,
"retardation_factor": 1.0,
"decay_rate": 0.0,
},
"ex-gwt-moc3d-p01c": {
"longitudinal_dispersivity": 1.0,
"retardation_factor": 2.0,
"decay_rate": 0.0,
},
"ex-gwt-moc3d-p01d": {
"longitudinal_dispersivity": 1.0,
"retardation_factor": 1.0,
"decay_rate": 0.01,
},
}
# Scenario parameter units - make sure there is at least one blank line before next item
# add parameter_units to add units to the scenario parameter table
parameter_units = {
"longitudinal_dispersivity": "$cm$",
"retardation_factor": "unitless",
"decay_rate": "$s^{-1}$",
}
# Model units
length_units = "centimeters"
time_units = "seconds"
# Model parameters
nper = 1 # Number of periods
nlay = 1 # Number of layers
nrow = 1 # Number of rows
ncol = 122 # Number of columns
system_length = 12.0 # Length of system ($cm$)
delr = 0.1 # Column width ($cm$)
delc = 0.1 # Row width ($cm$)
top = 1.0 # Top of the model ($cm$)
botm = 0 # Layer bottom elevation ($cm$)
specific_discharge = 0.1 # Specific discharge ($cm s^{-1}$)
hydraulic_conductivity = 0.01 # Hydraulic conductivity ($cm s^{-1}$)
porosity = 0.1 # Porosity of mobile domain (unitless)
total_time = 120.0 # Simulation time ($s$)
source_concentration = 1.0 # Source concentration (unitless)
initial_concentration = 0.0 # Initial concentration (unitless)
Model setup
Define functions to build models, write input files, and run the simulation.
[3]:
class Wexler1d:
"""
Analytical solution for 1D transport with inflow at a concentration of 1.
at x=0 and a third-type bound at location l.
Wexler Page 17 and Van Genuchten and Alves pages 66-67
"""
def betaeqn(self, beta, d, v, l):
return beta / np.tan(beta) - beta**2 * d / v / l + v * l / 4.0 / d
def fprimebetaeqn(self, beta, d, v, l):
"""
f1 = cotx - x/sinx2 - (2.0D0*C*x)
"""
c = v * l / 4.0 / d
return 1.0 / np.tan(beta) - beta / np.sin(beta) ** 2 - 2.0 * c * beta
def fprime2betaeqn(self, beta, d, v, l):
"""
f2 = -1.0D0/sinx2 - (sinx2-x*DSIN(x*2.0D0))/(sinx2*sinx2) - 2.0D0*C
"""
c = v * l / 4.0 / d
sinx2 = np.sin(beta) ** 2
return (
-1.0 / sinx2
- (sinx2 - beta * np.sin(beta * 2.0)) / (sinx2 * sinx2)
- 2.0 * c
)
def solvebetaeqn(self, beta, d, v, l, xtol=1.0e-12):
from scipy.optimize import fsolve
t = fsolve(
self.betaeqn,
beta,
args=(d, v, l),
fprime=self.fprime2betaeqn,
xtol=xtol,
full_output=True,
)
result = t[0][0]
infod = t[1]
isoln = t[2]
msg = t[3]
if abs(result - beta) > np.pi:
raise Exception("Error in beta solution")
err = self.betaeqn(result, d, v, l)
fvec = infod["fvec"][0]
if isoln != 1:
print("Error in beta solve", err, result, d, v, l, msg)
return result
def root3(self, d, v, l, nval=1000):
b = 0.5 * np.pi
betalist = []
for i in range(nval):
b = self.solvebetaeqn(b, d, v, l)
err = self.betaeqn(b, d, v, l)
betalist.append(b)
b += np.pi
return betalist
def analytical(self, x, t, v, l, d, tol=1.0e-20, nval=5000):
sigma = 0.0
betalist = self.root3(d, v, l, nval=nval)
concold = None
for i, bi in enumerate(betalist):
denom = bi**2 + (v * l / 2.0 / d) ** 2 + v * l / d
x1 = (
bi
* (bi * np.cos(bi * x / l) + v * l / 2.0 / d * np.sin(bi * x / l))
/ denom
)
denom = bi**2 + (v * l / 2.0 / d) ** 2
x2 = np.exp(-1 * bi**2 * d * t / l**2) / denom
sigma += x1 * x2
term1 = 2.0 * v * l / d * np.exp(v * x / 2.0 / d - v**2 * t / 4.0 / d)
conc = 1.0 - term1 * sigma
if i > 0:
assert concold is not None
diff = abs(conc - concold)
if np.all(diff < tol):
break
concold = conc
return conc
def analytical2(self, x, t, v, l, d, e=0.0, tol=1.0e-20, nval=5000):
"""
Calculate the analytical solution for one-dimension advection and
dispersion using the solution of Lapidus and Amundson (1952) and
Ogata and Banks (1961)
Parameters
----------
x : float or ndarray
x position
t : float or ndarray
time
v : float or ndarray
velocity
l : float
length domain
d : float
dispersion coefficient
e : float
decay rate
Returns
-------
result : float or ndarray
normalized concentration value
"""
u = v**2 + 4.0 * e * d
u = np.sqrt(u)
sigma = 0.0
denom = (u + v) / 2.0 / v - (u - v) ** 2.0 / 2.0 / v / (u + v) * np.exp(
-u * l / d
)
term1 = np.exp((v - u) * x / 2.0 / d) + (u - v) / (u + v) * np.exp(
(v + u) * x / 2.0 / d - u * l / d
)
term1 = term1 / denom
term2 = 2.0 * v * l / d * np.exp(v * x / 2.0 / d - v**2 * t / 4.0 / d - e * t)
betalist = self.root3(d, v, l, nval=nval)
concold = None
for i, bi in enumerate(betalist):
denom = bi**2 + (v * l / 2.0 / d) ** 2 + v * l / d
x1 = (
bi
* (bi * np.cos(bi * x / l) + v * l / 2.0 / d * np.sin(bi * x / l))
/ denom
)
denom = bi**2 + (v * l / 2.0 / d) ** 2 + e * l**2 / d
x2 = np.exp(-1 * bi**2 * d * t / l**2) / denom
sigma += x1 * x2
conc = term1 - term2 * sigma
if i > 0:
assert concold is not None
diff = abs(conc - concold)
if np.all(diff < tol):
break
concold = conc
return conc
def get_sorption_dict(retardation_factor):
sorption = None
bulk_density = None
distcoef = None
if retardation_factor > 1.0:
sorption = "linear"
bulk_density = 1.0
distcoef = (retardation_factor - 1.0) * porosity / bulk_density
sorption_dict = {
"sorption": sorption,
"bulk_density": bulk_density,
"distcoef": distcoef,
}
return sorption_dict
def get_decay_dict(decay_rate, sorption=False):
first_order_decay = None
decay = None
decay_sorbed = None
if decay_rate != 0.0:
first_order_decay = True
decay = decay_rate
if sorption:
decay_sorbed = decay_rate
decay_dict = {
"first_order_decay": first_order_decay,
"decay": decay,
"decay_sorbed": decay_sorbed,
}
return decay_dict
def build_mf6gwf(sim_folder):
print(f"Building mf6gwf model...{sim_folder}")
name = "flow"
sim_ws = os.path.join(workspace, sim_folder, "mf6gwf")
sim = flopy.mf6.MFSimulation(sim_name=name, sim_ws=sim_ws, exe_name="mf6")
tdis_ds = ((total_time, 1, 1.0),)
flopy.mf6.ModflowTdis(sim, nper=nper, perioddata=tdis_ds, time_units=time_units)
flopy.mf6.ModflowIms(sim)
gwf = flopy.mf6.ModflowGwf(sim, modelname=name, save_flows=True)
flopy.mf6.ModflowGwfdis(
gwf,
length_units=length_units,
nlay=nlay,
nrow=nrow,
ncol=ncol,
delr=delr,
delc=delc,
top=top,
botm=botm,
)
flopy.mf6.ModflowGwfnpf(
gwf,
save_specific_discharge=True,
save_saturation=True,
icelltype=0,
k=hydraulic_conductivity,
)
flopy.mf6.ModflowGwfic(gwf, strt=1.0)
flopy.mf6.ModflowGwfchd(gwf, stress_period_data=[[(0, 0, ncol - 1), 1.0]])
wel_spd = {
0: [
[
(0, 0, 0),
specific_discharge * delc * delr * top,
source_concentration,
]
],
}
flopy.mf6.ModflowGwfwel(
gwf,
stress_period_data=wel_spd,
pname="WEL-1",
auxiliary=["CONCENTRATION"],
)
head_filerecord = f"{name}.hds"
budget_filerecord = f"{name}.bud"
flopy.mf6.ModflowGwfoc(
gwf,
head_filerecord=head_filerecord,
budget_filerecord=budget_filerecord,
saverecord=[("HEAD", "ALL"), ("BUDGET", "ALL")],
)
return sim
def build_mf6gwt(sim_folder, longitudinal_dispersivity, retardation_factor, decay_rate):
print(f"Building mf6gwt model...{sim_folder}")
name = "trans"
sim_ws = os.path.join(workspace, sim_folder, "mf6gwt")
sim = flopy.mf6.MFSimulation(sim_name=name, sim_ws=sim_ws, exe_name="mf6")
tdis_ds = ((total_time, 240, 1.0),)
flopy.mf6.ModflowTdis(sim, nper=nper, perioddata=tdis_ds, time_units=time_units)
flopy.mf6.ModflowIms(sim, linear_acceleration="bicgstab")
gwt = flopy.mf6.ModflowGwt(sim, modelname=name, save_flows=True)
flopy.mf6.ModflowGwtdis(
gwt,
length_units=length_units,
nlay=nlay,
nrow=nrow,
ncol=ncol,
delr=delr,
delc=delc,
top=top,
botm=botm,
)
flopy.mf6.ModflowGwtic(gwt, strt=0)
flopy.mf6.ModflowGwtmst(
gwt,
porosity=porosity,
**get_sorption_dict(retardation_factor),
**get_decay_dict(decay_rate, retardation_factor > 1.0),
)
flopy.mf6.ModflowGwtadv(gwt, scheme="TVD")
flopy.mf6.ModflowGwtdsp(
gwt,
xt3d_off=True,
alh=longitudinal_dispersivity,
ath1=longitudinal_dispersivity,
)
pd = [
("GWFHEAD", "../mf6gwf/flow.hds", None),
("GWFBUDGET", "../mf6gwf/flow.bud", None),
]
flopy.mf6.ModflowGwtfmi(gwt, packagedata=pd)
sourcerecarray = [["WEL-1", "AUX", "CONCENTRATION"]]
flopy.mf6.ModflowGwtssm(gwt, sources=sourcerecarray)
# flopy.mf6.ModflowGwtcnc(gwt, stress_period_data=[((0, 0, 0), source_concentration),])
obs_data = {
f"{name}.obs.csv": [
("X005", "CONCENTRATION", (0, 0, 0)),
("X405", "CONCENTRATION", (0, 0, 40)),
("X1105", "CONCENTRATION", (0, 0, 110)),
],
}
obs_package = flopy.mf6.ModflowUtlobs(
gwt, digits=10, print_input=True, continuous=obs_data
)
flopy.mf6.ModflowGwtoc(
gwt,
budget_filerecord=f"{name}.cbc",
concentration_filerecord=f"{name}.ucn",
saverecord=[("CONCENTRATION", "ALL"), ("BUDGET", "LAST")],
printrecord=[("CONCENTRATION", "LAST"), ("BUDGET", "LAST")],
)
return sim
def build_models(sim_name, longitudinal_dispersivity, retardation_factor, decay_rate):
sim_mf6gwf = build_mf6gwf(sim_name)
sim_mf6gwt = build_mf6gwt(
sim_name, longitudinal_dispersivity, retardation_factor, decay_rate
)
return sim_mf6gwf, sim_mf6gwt
def write_models(sims, silent=True):
sim_mf6gwf, sim_mf6gwt = sims
sim_mf6gwf.write_simulation(silent=silent)
sim_mf6gwt.write_simulation(silent=silent)
@timed
def run_models(sims, silent=True):
sim_mf6gwf, sim_mf6gwt = sims
success, buff = sim_mf6gwf.run_simulation(silent=silent)
assert success, buff
success, buff = sim_mf6gwt.run_simulation(silent=silent)
assert success, buff
Plotting results
Define functions to plot model results.
[4]:
# Figure properties
figure_size = (5, 3)
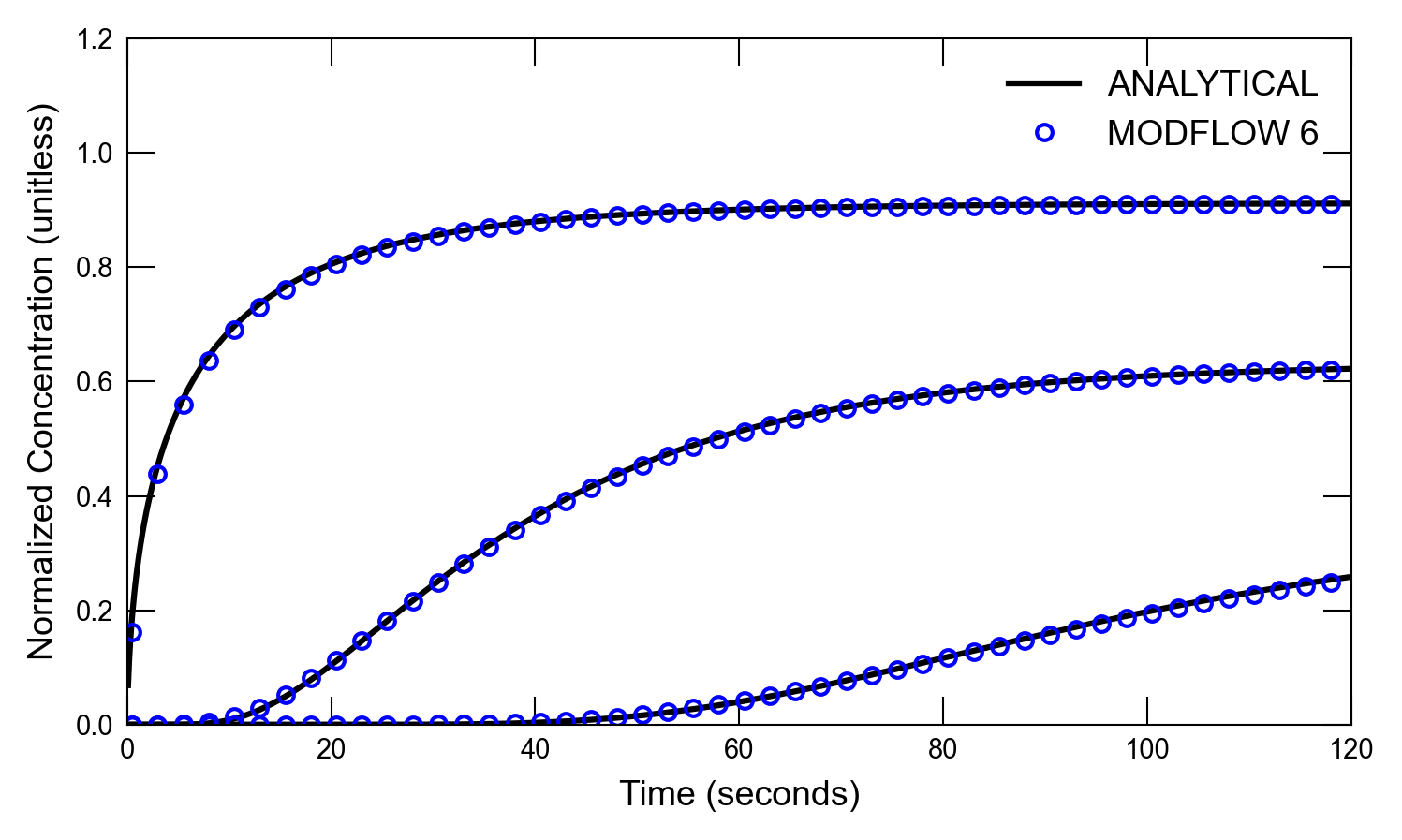
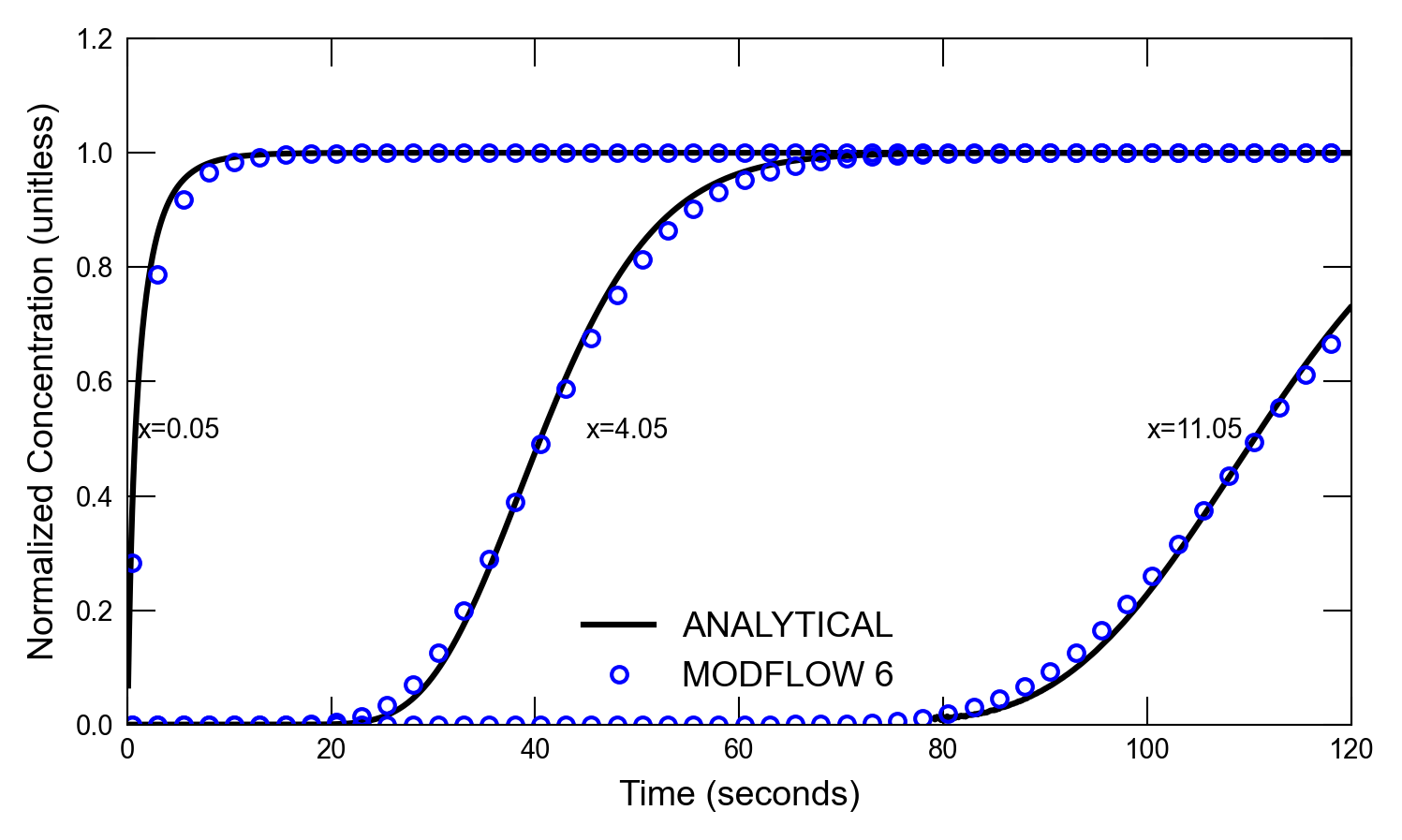
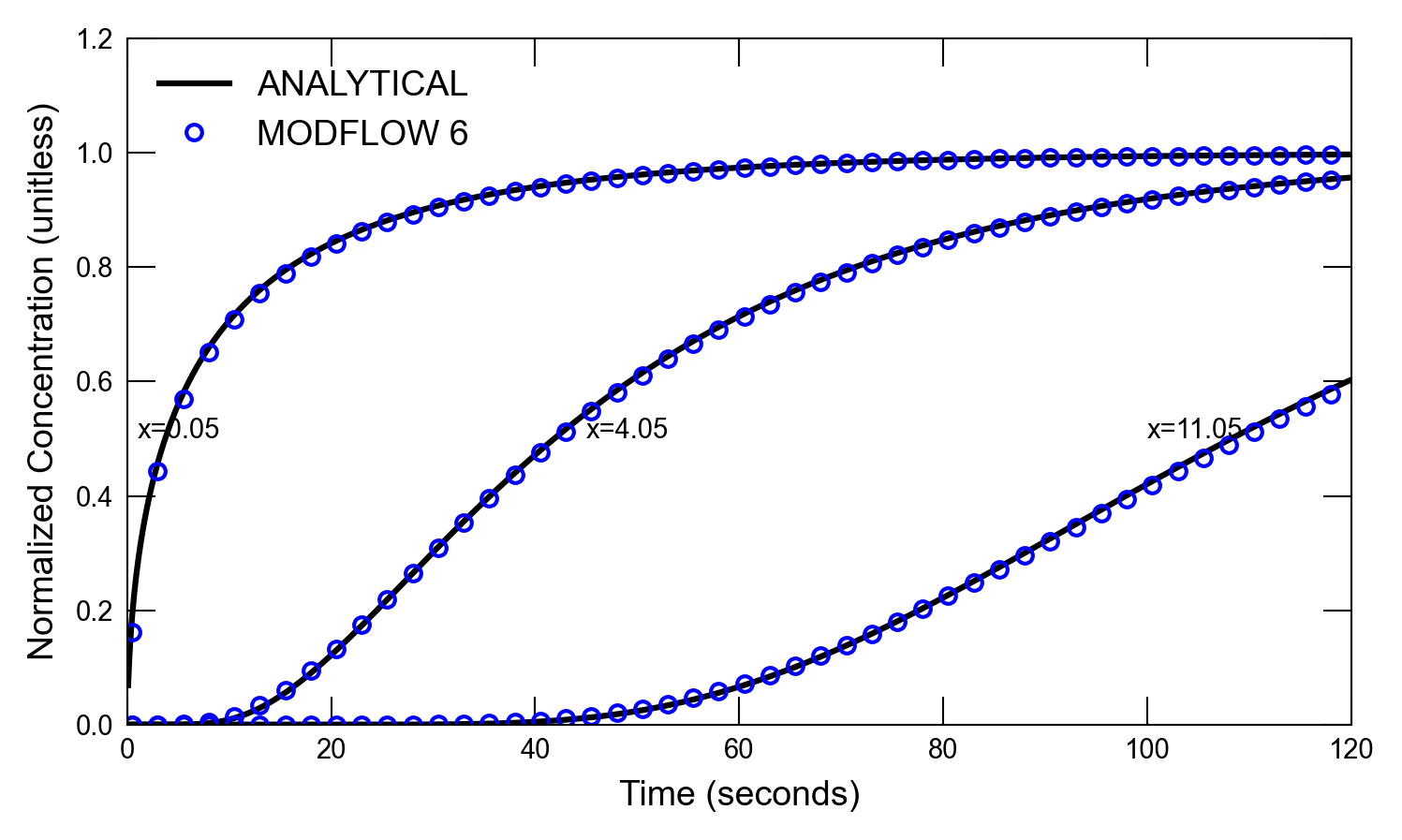
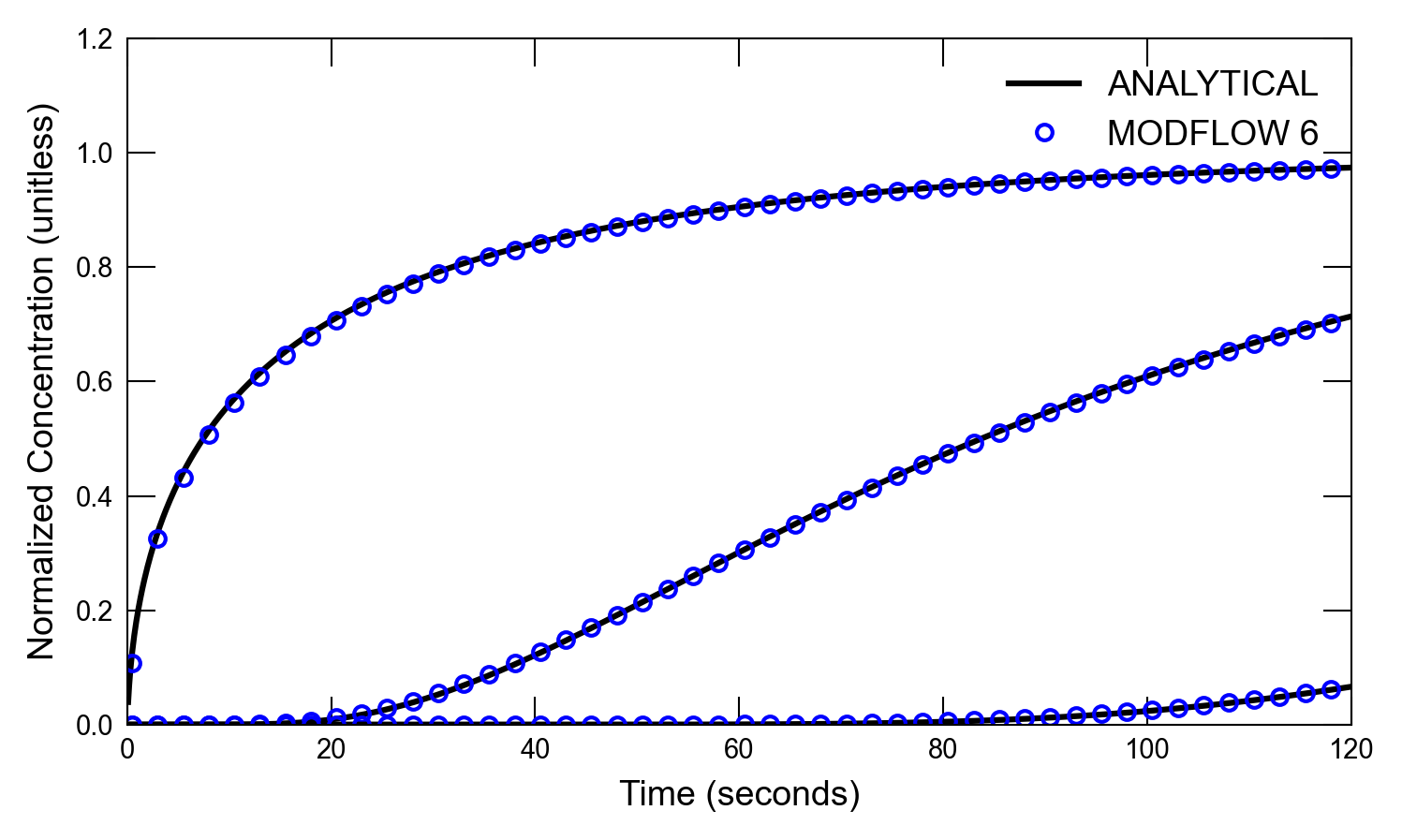
def plot_results_ct(
sims, idx, longitudinal_dispersivity, retardation_factor, decay_rate
):
_, sim_mf6gwt = sims
with styles.USGSPlot():
sim_ws = sim_mf6gwt.simulation_data.mfpath.get_sim_path()
mf6gwt_ra = sim_mf6gwt.get_model("trans").obs.output.obs().data
fig, axs = plt.subplots(1, 1, figsize=figure_size, dpi=300, tight_layout=True)
alabel = ["ANALYTICAL", "", ""]
mlabel = ["MODFLOW 6", "", ""]
iskip = 5
atimes = np.arange(0, total_time, 0.1)
obsnames = ["X005", "X405", "X1105"]
simtimes = mf6gwt_ra["totim"]
dispersion_coefficient = (
longitudinal_dispersivity * specific_discharge / retardation_factor
)
for i, x in enumerate([0.05, 4.05, 11.05]):
a1 = Wexler1d().analytical2(
x,
atimes,
specific_discharge / retardation_factor,
system_length,
dispersion_coefficient,
decay_rate,
)
if idx == 0:
idx_filter = a1 < 0
a1[idx_filter] = 0
idx_filter = a1 > 1
a1[idx_filter] = 0
idx_filter = atimes > 0
if i == 2:
idx_filter = atimes > 79
elif idx > 0:
idx_filter = atimes > 0
axs.plot(atimes[idx_filter], a1[idx_filter], color="k", label=alabel[i])
axs.plot(
simtimes[::iskip],
mf6gwt_ra[obsnames[i]][::iskip],
marker="o",
ls="none",
mec="blue",
mfc="none",
markersize="4",
label=mlabel[i],
)
axs.set_ylim(0, 1.2)
axs.set_xlim(0, 120)
axs.set_xlabel("Time (seconds)")
axs.set_ylabel("Normalized Concentration (unitless)")
if idx in [0, 1]:
axs.text(1, 0.5, "x=0.05")
axs.text(45, 0.5, "x=4.05")
axs.text(100, 0.5, "x=11.05")
axs.legend()
if plot_show:
plt.show()
if plot_save:
sim_folder = os.path.split(sim_ws)[0]
sim_folder = os.path.basename(sim_folder)
fname = f"{sim_folder}-ct.png"
fpth = figs_path / fname
fig.savefig(fpth)
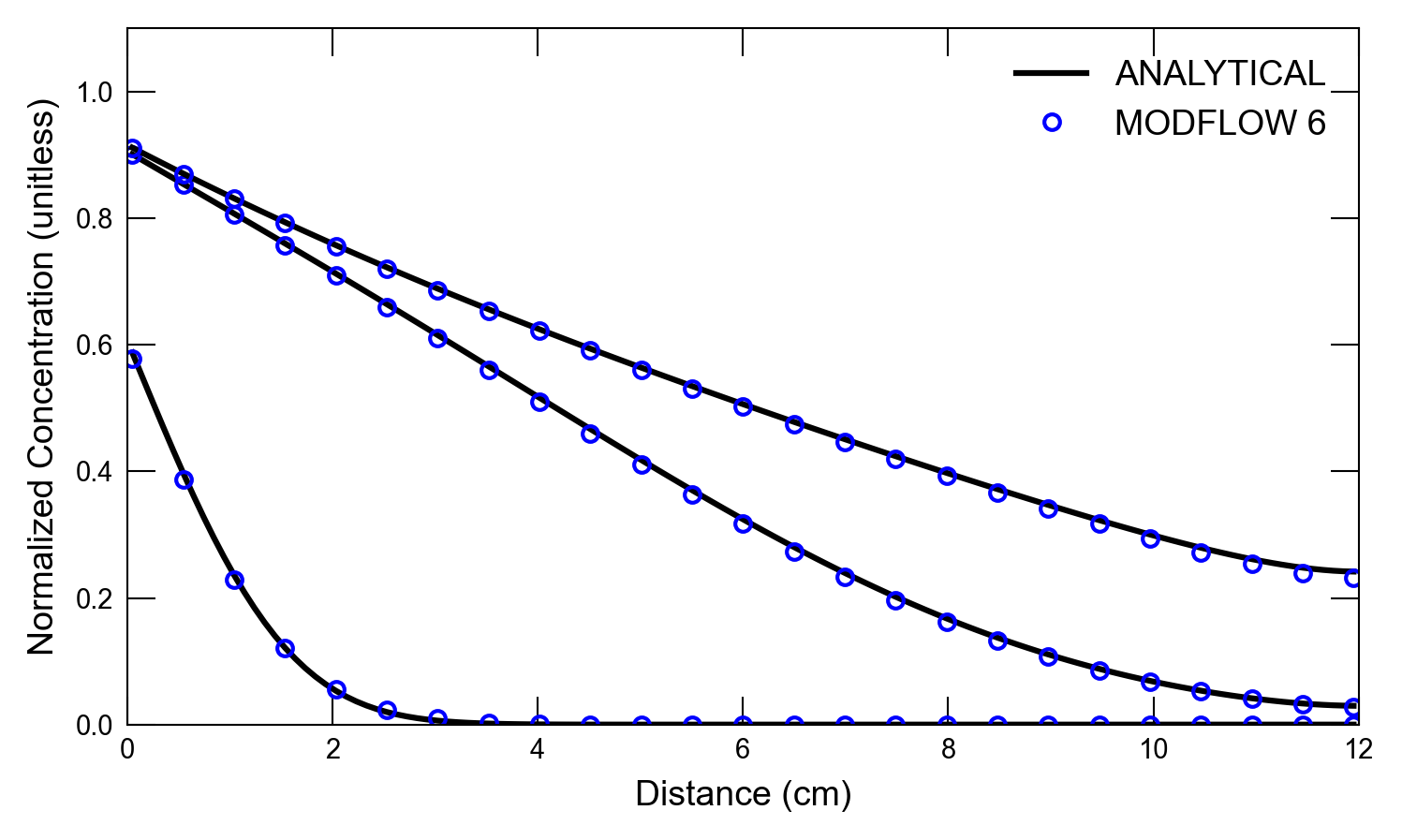
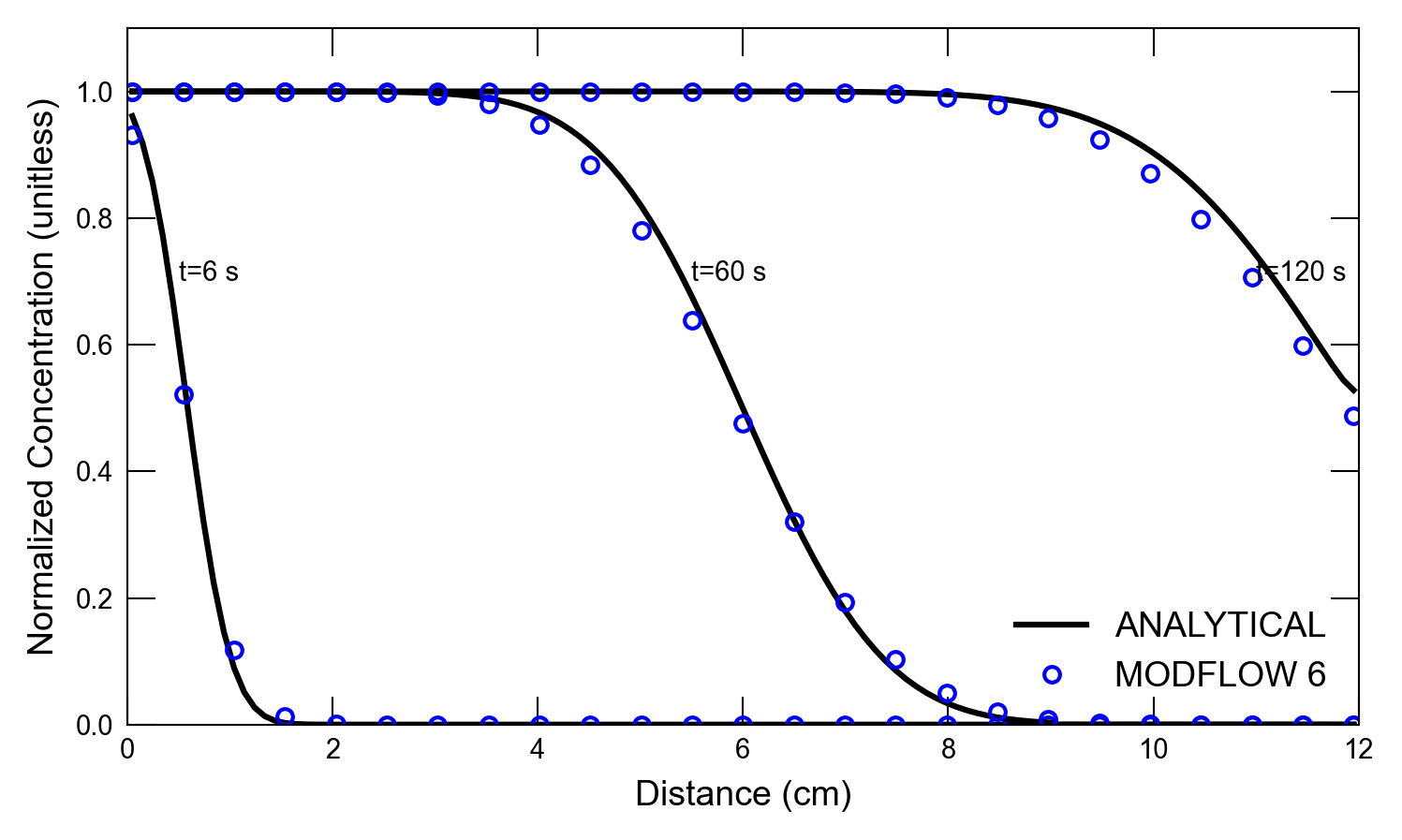
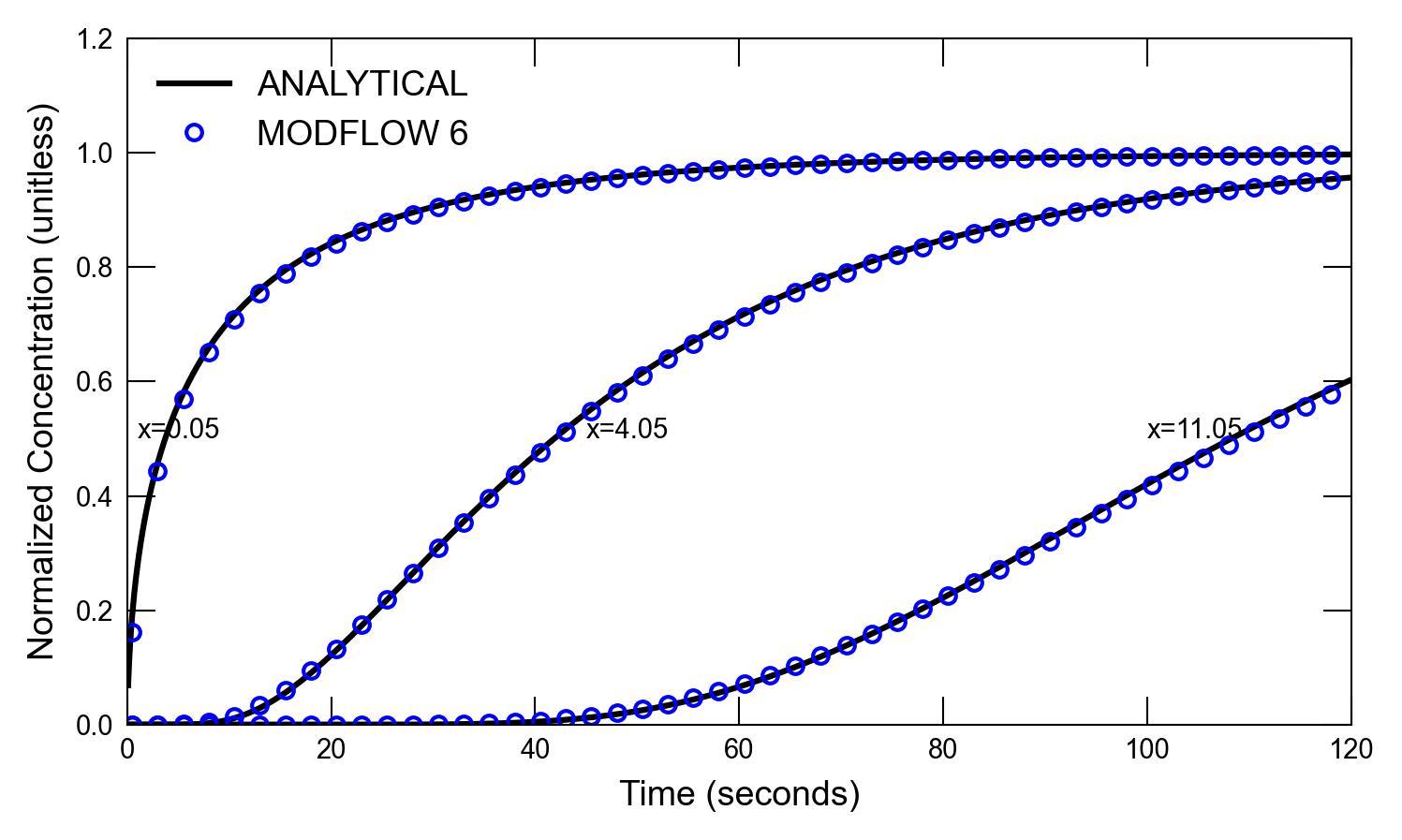
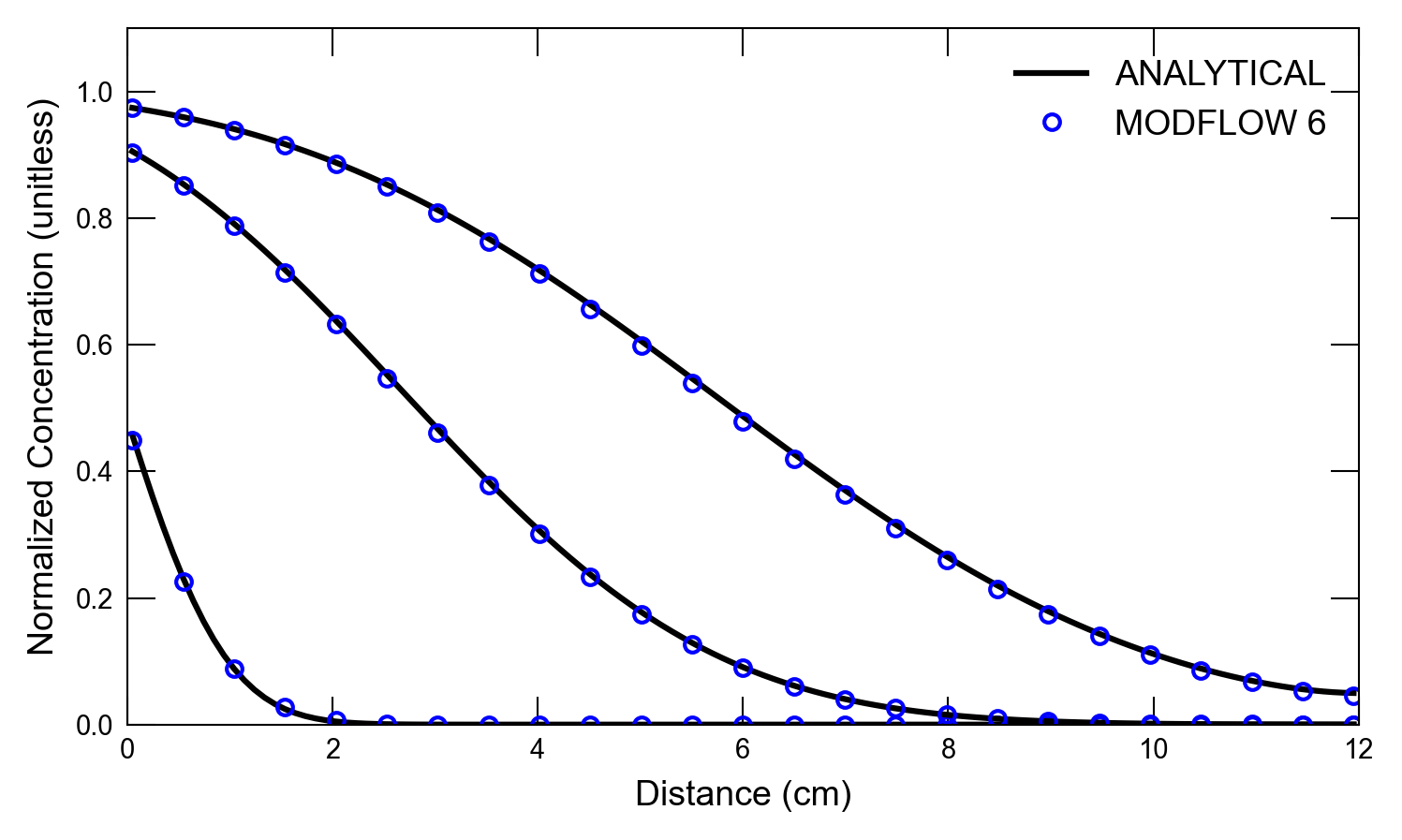
def plot_results_cd(
sims, idx, longitudinal_dispersivity, retardation_factor, decay_rate
):
_, sim_mf6gwt = sims
with styles.USGSPlot():
ucnobj_mf6 = sim_mf6gwt.trans.output.concentration()
fig, axs = plt.subplots(1, 1, figsize=figure_size, dpi=300, tight_layout=True)
alabel = ["ANALYTICAL", "", ""]
mlabel = ["MODFLOW 6", "", ""]
iskip = 5
ctimes = [6.0, 60.0, 120.0]
x = np.linspace(0.5 * delr, system_length - 0.5 * delr, ncol - 1)
dispersion_coefficient = (
longitudinal_dispersivity * specific_discharge / retardation_factor
)
for i, t in enumerate(ctimes):
a1 = Wexler1d().analytical2(
x,
t,
specific_discharge / retardation_factor,
system_length,
dispersion_coefficient,
decay_rate,
)
if idx == 0:
idx_filter = x > system_length
if i == 0:
idx_filter = x > 6
if i == 1:
idx_filter = x > 9
a1[idx_filter] = 0.0
axs.plot(x, a1, color="k", label=alabel[i])
simconc = ucnobj_mf6.get_data(totim=t).flatten()
axs.plot(
x[::iskip],
simconc[::iskip],
marker="o",
ls="none",
mec="blue",
mfc="none",
markersize="4",
label=mlabel[i],
)
axs.set_ylim(0, 1.1)
axs.set_xlim(0, 12)
if idx in [0, 1]:
axs.text(0.5, 0.7, "t=6 s")
axs.text(5.5, 0.7, "t=60 s")
axs.text(11, 0.7, "t=120 s")
axs.set_xlabel("Distance (cm)")
axs.set_ylabel("Normalized Concentration (unitless)")
plt.legend()
if plot_show:
plt.show()
if plot_save:
sim_ws = sim_mf6gwt.simulation_data.mfpath.get_sim_path()
sim_folder = os.path.split(sim_ws)[0]
sim_folder = os.path.basename(sim_folder)
fname = f"{sim_folder}-cd.png"
fpth = figs_path / fname
fig.savefig(fpth)
Running the example
Define and invoke a function to run the example scenario, then plot results.
[5]:
def scenario(idx, silent=True):
key = list(parameters.keys())[idx]
parameter_dict = parameters[key]
sims = build_models(key, **parameter_dict)
if write:
write_models(sims, silent=silent)
if run:
run_models(sims, silent=silent)
if plot:
plot_results_ct(sims, idx, **parameter_dict)
plot_results_cd(sims, idx, **parameter_dict)
[6]:
scenario(0)
Building mf6gwf model...ex-gwt-moc3d-p01a
Building mf6gwt model...ex-gwt-moc3d-p01a
run_models took 127.79 ms
[7]:
scenario(1)
Building mf6gwf model...ex-gwt-moc3d-p01b
Building mf6gwt model...ex-gwt-moc3d-p01b
run_models took 130.28 ms
[8]:
scenario(2)
Building mf6gwf model...ex-gwt-moc3d-p01c
Building mf6gwt model...ex-gwt-moc3d-p01c
run_models took 171.13 ms
[9]:
scenario(3)
Building mf6gwf model...ex-gwt-moc3d-p01d
Building mf6gwt model...ex-gwt-moc3d-p01d
run_models took 128.94 ms