This page was generated from
ex-gwf-csub-p04.py.
It's also available as a notebook.
One-dimensional compaction in a three-dimensional flow field example
This problem is based on the problem presented in the SUB-WT report (Leake and Galloway, 2007) and represent groundwater development in a hypothetical aquifer that includes some features typical of basin-fill aquifers in an arid or semi-arid environment.
Initial setup
Import dependencies, define the example name and workspace, and read settings from environment variables.
[1]:
import os
import pathlib as pl
import flopy
import git
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pooch
from flopy.plot.styles import styles
from modflow_devtools.misc import get_env, timed
# Example name and workspace paths. If this example is running
# in the git repository, use the folder structure described in
# the README. Otherwise just use the current working directory.
sim_name = "ex-gwf-csub-p04"
try:
root = pl.Path(git.Repo(".", search_parent_directories=True).working_dir)
except:
root = None
workspace = root / "examples" if root else pl.Path.cwd()
figs_path = root / "figures" if root else pl.Path.cwd()
data_path = root / "data" / sim_name if root else pl.Path.cwd()
# Settings from environment variables
write = get_env("WRITE", True)
run = get_env("RUN", True)
plot = get_env("PLOT", True)
plot_show = get_env("PLOT_SHOW", True)
plot_save = get_env("PLOT_SAVE", True)
Define parameters
Define model units, parameters and other settings.
[2]:
# Model units
length_units = "meters"
time_units = "days"
# Model parameters
nper = 3 # Number of periods
nlay = 4 # Number of layers
nrow = 20 # Number of rows
ncol = 15 # Number of columns
delr = 2000.0 # Column width ($m$)
delc = 2000.0 # Row width ($m$)
top = 150.0 # Top of the model ($ft$)
botm_str = "50., -100., -150., -350." # Layer bottom elevations ($m$)
strt = 100.0 # Starting head ($m$)
icelltype_str = "1, 0, 0, 0" # Cell conversion type
k11_str = "4., 4., 0.01, 4." # Horizontal hydraulic conductivity ($m/d$)
k33_str = "0.4, 0.4, 0.01, 0.4" # Vertical hydraulic conductivity ($m/d$)
sy_str = "0.3, 0.3, 0.4, 0.3" # Specific yield (unitless)
gammaw = 9806.65 # Compressibility of water (Newtons/($m^3$)
beta = 4.6612e-10 # Specific gravity of water (1/$Pa$)
sgm_str = "1.77, 1.77, 1.60, 1.77" # Specific gravity of moist soils (unitless)
sgs_str = "2.06, 2.05, 1.94, 2.06" # Specific gravity of saturated soils (unitless)
cg_theta_str = "0.32, 0.32, 0.45, 0.32" # Coarse-grained material porosity (unitless)
cg_ske_str = "0.005, 0.005, 0.01, 0.005" # Elastic specific storage ($1/m$)
ib_thick_str = "45., 70., 50., 90." # Interbed thickness ($m$)
ib_theta = 0.45 # Interbed initial porosity (unitless)
ib_cr = 0.01 # Interbed recompression index (unitless)
ib_cv = 0.25 # Interbed compression index (unitless)
stress_offset = 15.0 # Initial preconsolidation stress offset ($m$)
# Static temporal data used by TDIS file
tdis_ds = (
(0.0, 1, 1.0),
(21915.0, 60, 1.0),
(21915.0, 60, 1.0),
)
# Parse parameter strings into tuples
botm = [float(value) for value in botm_str.split(",")]
icelltype = [int(value) for value in icelltype_str.split(",")]
k11 = [float(value) for value in k11_str.split(",")]
k33 = [float(value) for value in k33_str.split(",")]
sy = [float(value) for value in sy_str.split(",")]
sgm = [float(value) for value in sgm_str.split(",")]
sgs = [float(value) for value in sgs_str.split(",")]
cg_theta = [float(value) for value in cg_theta_str.split(",")]
cg_ske = [float(value) for value in cg_ske_str.split(",")]
ib_thick = [float(value) for value in ib_thick_str.split(",")]
# Load active domain and create idomain array
fname = "idomain.txt"
fpath = pooch.retrieve(
url=f"https://github.com/MODFLOW-USGS/modflow6-examples/raw/master/data/{sim_name}/{fname}",
fname=fname,
path=data_path,
known_hash="md5:2f05a27b6f71e564c0d3616e3fd00ac8",
)
ib = np.loadtxt(fpath, dtype=int)
idomain = np.tile(ib, (nlay, 1))
# Constant head boundary cells
chd_locs = [(nrow - 1, 7), (nrow - 1, 8)]
c6 = []
for i, j in chd_locs:
for k in range(nlay):
c6.append([k, i, j, strt])
# Recharge boundary cells
rch_rate = 5.5e-4
rch6 = []
for i in range(nrow):
for j in range(ncol):
if ib[i, j] != 2 or (i, j) in chd_locs:
continue
rch6.append([0, i, j, rch_rate])
# Well boundary cells
well_locs = (
(1, 8, 9),
(3, 11, 6),
)
well_rates = (
-72000,
0.0,
)
wel6 = {}
for idx, q in enumerate(well_rates):
spd = []
for k, i, j in well_locs:
spd.append([k, i, j, q])
wel6[idx + 1] = spd
# Create interbed package data
icsubno = 0
csub_pakdata = []
for i in range(nrow):
for j in range(ncol):
if ib[i, j] < 1 or (i, j) in chd_locs:
continue
for k in range(nlay):
boundname = f"{k + 1:02d}_{i + 1:02d}_{j + 1:02d}"
ib_lst = [
icsubno,
(k, i, j),
"nodelay",
stress_offset,
ib_thick[k],
1.0,
ib_cv,
ib_cr,
ib_theta,
999.0,
999.0,
boundname,
]
csub_pakdata.append(ib_lst)
icsubno += 1
# Solver parameters
nouter = 100
ninner = 300
hclose = 1e-9
rclose = 1e-6
linaccel = "bicgstab"
relax = 0.97
Model setup
Define functions to build models, write input files, and run the simulation.
[3]:
def build_models():
sim_ws = os.path.join(workspace, sim_name)
sim = flopy.mf6.MFSimulation(sim_name=sim_name, sim_ws=sim_ws, exe_name="mf6")
flopy.mf6.ModflowTdis(sim, nper=nper, perioddata=tdis_ds, time_units=time_units)
flopy.mf6.ModflowIms(
sim,
outer_maximum=nouter,
outer_dvclose=hclose,
linear_acceleration=linaccel,
inner_maximum=ninner,
inner_dvclose=hclose,
relaxation_factor=relax,
rcloserecord=f"{rclose} strict",
)
gwf = flopy.mf6.ModflowGwf(
sim, modelname=sim_name, save_flows=True, newtonoptions="newton"
)
flopy.mf6.ModflowGwfdis(
gwf,
length_units=length_units,
nlay=nlay,
nrow=nrow,
ncol=ncol,
delr=delr,
delc=delc,
top=top,
botm=botm,
idomain=idomain,
)
# gwf obs
flopy.mf6.ModflowUtlobs(
gwf,
digits=10,
print_input=True,
continuous={
"gwf_obs.csv": [
("h1l1", "HEAD", (0, 8, 9)),
("h1l2", "HEAD", (1, 8, 9)),
("h1l3", "HEAD", (2, 8, 9)),
("h1l4", "HEAD", (3, 8, 9)),
("h2l1", "HEAD", (0, 11, 6)),
("h2l2", "HEAD", (1, 11, 6)),
("h3l2", "HEAD", (2, 11, 6)),
("h4l2", "HEAD", (3, 11, 6)),
]
},
)
flopy.mf6.ModflowGwfic(gwf, strt=strt)
flopy.mf6.ModflowGwfnpf(
gwf,
icelltype=icelltype,
k=k11,
save_specific_discharge=True,
)
flopy.mf6.ModflowGwfsto(
gwf,
iconvert=icelltype,
ss=0.0,
sy=sy,
steady_state={0: True},
transient={1: True},
)
csub = flopy.mf6.ModflowGwfcsub(
gwf,
print_input=True,
save_flows=True,
compression_indices=True,
update_material_properties=True,
boundnames=True,
ninterbeds=len(csub_pakdata),
sgm=sgm,
sgs=sgs,
cg_theta=cg_theta,
cg_ske_cr=cg_ske,
beta=beta,
gammaw=gammaw,
packagedata=csub_pakdata,
)
opth = f"{sim_name}.csub.obs"
csub_csv = opth + ".csv"
obs = [
("w1l1", "interbed-compaction", "01_09_10"),
("w1l2", "interbed-compaction", "02_09_10"),
("w1l3", "interbed-compaction", "03_09_10"),
("w1l4", "interbed-compaction", "04_09_10"),
("w2l1", "interbed-compaction", "01_12_07"),
("w2l2", "interbed-compaction", "02_12_07"),
("w2l3", "interbed-compaction", "03_12_07"),
("w2l4", "interbed-compaction", "04_12_07"),
("s1l1", "coarse-compaction", (0, 8, 9)),
("s1l2", "coarse-compaction", (1, 8, 9)),
("s1l3", "coarse-compaction", (2, 8, 9)),
("s1l4", "coarse-compaction", (3, 8, 9)),
("s2l1", "coarse-compaction", (0, 11, 6)),
("s2l2", "coarse-compaction", (1, 11, 6)),
("s2l3", "coarse-compaction", (2, 11, 6)),
("s2l4", "coarse-compaction", (3, 11, 6)),
("c1l1", "compaction-cell", (0, 8, 9)),
("c1l2", "compaction-cell", (1, 8, 9)),
("c1l3", "compaction-cell", (2, 8, 9)),
("c1l4", "compaction-cell", (3, 8, 9)),
("c2l1", "compaction-cell", (0, 11, 6)),
("c2l2", "compaction-cell", (1, 11, 6)),
("c2l3", "compaction-cell", (2, 11, 6)),
("c2l4", "compaction-cell", (3, 11, 6)),
("w2l4q", "csub-cell", (3, 11, 6)),
("gs1", "gstress-cell", (0, 8, 9)),
("es1", "estress-cell", (0, 8, 9)),
("pc1", "preconstress-cell", (0, 8, 9)),
("gs2", "gstress-cell", (1, 8, 9)),
("es2", "estress-cell", (1, 8, 9)),
("pc2", "preconstress-cell", (1, 8, 9)),
("gs3", "gstress-cell", (2, 8, 9)),
("es3", "estress-cell", (2, 8, 9)),
("pc3", "preconstress-cell", (2, 8, 9)),
("gs4", "gstress-cell", (3, 8, 9)),
("es4", "estress-cell", (3, 8, 9)),
("pc4", "preconstress-cell", (3, 8, 9)),
("sk1l2", "ske-cell", (1, 8, 9)),
("sk2l4", "ske-cell", (3, 11, 6)),
("t1l2", "theta", "02_09_10"),
("w1qie", "elastic-csub", "02_09_10"),
("w1qii", "inelastic-csub", "02_09_10"),
("w1qaq", "coarse-csub", (1, 8, 9)),
("w1qt", "csub-cell", (1, 8, 9)),
("w1wc", "wcomp-csub-cell", (1, 8, 9)),
("w2qie", "elastic-csub", "04_12_07"),
("w2qii", "inelastic-csub", "04_12_07"),
("w2qaq", "coarse-csub", (3, 11, 6)),
("w2qt ", "csub-cell", (3, 11, 6)),
("w2wc", "wcomp-csub-cell", (3, 11, 6)),
]
orecarray = {csub_csv: obs}
csub.obs.initialize(
filename=opth, digits=10, print_input=True, continuous=orecarray
)
flopy.mf6.ModflowGwfchd(gwf, stress_period_data={0: c6})
flopy.mf6.ModflowGwfrch(gwf, stress_period_data={0: rch6})
flopy.mf6.ModflowGwfwel(gwf, stress_period_data=wel6)
head_filerecord = f"{sim_name}.hds"
budget_filerecord = f"{sim_name}.cbc"
flopy.mf6.ModflowGwfoc(
gwf,
head_filerecord=head_filerecord,
budget_filerecord=budget_filerecord,
printrecord=[("BUDGET", "ALL")],
saverecord=[("BUDGET", "ALL"), ("HEAD", "ALL")],
)
return sim
def write_models(sim, silent=True):
sim.write_simulation(silent=silent)
@timed
def run_models(sim, silent=True):
success, buff = sim.run_simulation(silent=silent)
assert success, buff
Plotting results
Define functions to plot model results, starting with a few utilities.
[4]:
# Set figure properties specific to the problem
figure_size = (6.8, 5.5)
arrow_props = dict(facecolor="black", arrowstyle="-", lw=0.5)
plot_tags = (
"W1L",
"W2L",
"S1L",
"S2L",
"C1L",
"C2L",
)
compaction_heading = ("row 9, column 10", "row 12, column 7")
def get_csub_observations(sim):
name = sim.name
gwf = sim.get_model(sim_name)
csub_obs = gwf.csub.output.obs().data
csub_obs["totim"] /= 365.25
# set initial preconsolidation stress to stress period 1 value
slist = [name for name in csub_obs.dtype.names if "PC" in name]
for tag in slist:
csub_obs[tag][0] = csub_obs[tag][1]
# set initial storativity to stress period 1 value
sk_tags = (
"SK1L2",
"SK2L4",
)
for tag in sk_tags:
if tag in csub_obs.dtype.names:
csub_obs[tag][0] = csub_obs[tag][1]
return csub_obs
def calc_compaction_at_surface(sim):
"""Calculate the compaction at the surface"""
csub_obs = get_csub_observations(sim)
for tag in plot_tags:
for k in (
3,
2,
1,
):
tag0 = f"{tag}{k}"
tag1 = f"{tag}{k + 1}"
csub_obs[tag0] += csub_obs[tag1]
return csub_obs
def plot_compaction_values(ax, sim, tagbase="W1L"):
colors = ["#FFF8DC", "#D2B48C", "#CD853F", "#8B4513"][::-1]
obs = calc_compaction_at_surface(sim)
for k in range(nlay):
fc = colors[k]
tag = f"{tagbase}{k + 1}"
label = f"Layer {k + 1}"
ax.fill_between(obs["totim"], obs[tag], y2=0, color=fc, label=label)
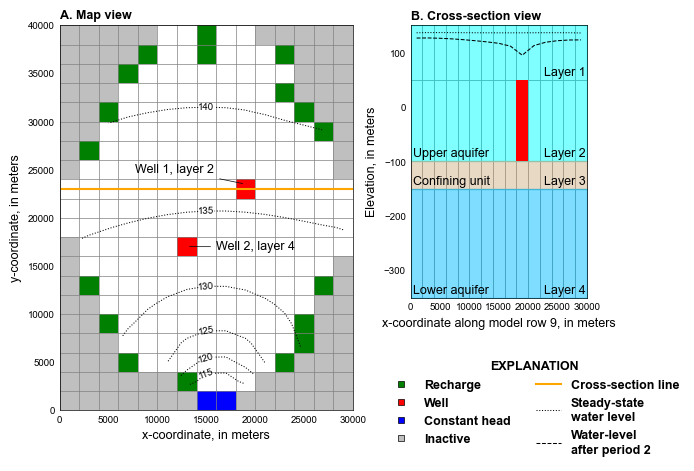
def plot_grid(sim, silent=True):
with styles.USGSMap():
name = sim.name
gwf = sim.get_model(name)
extents = gwf.modelgrid.extent
# read simulated heads
hobj = gwf.output.head()
h0 = hobj.get_data(kstpkper=(0, 0))
h1 = hobj.get_data(kstpkper=(59, 1))
hsxs0 = h0[0, 8, :]
hsxs1 = h1[0, 8, :]
# get delr array
dx = gwf.dis.delr.array
# create x-axis for cross-section
hxloc = np.arange(1000, 2000.0 * 15, 2000.0)
# set cross-section location
y = 2000.0 * 11.5
xsloc = [(extents[0], extents[1]), (y, y)]
# well locations
w1loc = (9.5 * 2000.0, 11.75 * 2000.0)
w2loc = (6.5 * 2000.0, 8.5 * 2000.0)
fig = plt.figure(figsize=(6.8, 5), constrained_layout=True)
gs = mpl.gridspec.GridSpec(7, 10, figure=fig, wspace=100)
plt.axis("off")
ax = fig.add_subplot(gs[:, 0:6])
# ax.set_aspect('equal')
mm = flopy.plot.PlotMapView(model=gwf, ax=ax, extent=extents)
mm.plot_grid(lw=0.5, color="0.5")
mm.plot_bc(ftype="WEL", kper=1, plotAll=True)
mm.plot_bc(ftype="CHD", color="blue")
mm.plot_bc(ftype="RCH", color="green")
mm.plot_inactive(color_noflow="0.75")
mm.ax.plot(xsloc[0], xsloc[1], color="orange", lw=1.5)
# contour steady state heads
cl = mm.contour_array(
h0,
masked_values=[1.0e30],
levels=np.arange(115, 200, 5),
colors="black",
linestyles="dotted",
linewidths=0.75,
)
ax.clabel(cl, fmt="%3i", inline_spacing=0.1)
# well text
styles.add_annotation(
ax=ax,
text="Well 1, layer 2",
bold=False,
italic=False,
xy=w1loc,
xytext=(w1loc[0] - 3200, w1loc[1] + 1500),
ha="right",
va="center",
zorder=100,
arrowprops=arrow_props,
)
styles.add_annotation(
ax=ax,
text="Well 2, layer 4",
bold=False,
italic=False,
xy=w2loc,
xytext=(w2loc[0] + 3000, w2loc[1]),
ha="left",
va="center",
zorder=100,
arrowprops=arrow_props,
)
ax.set_ylabel("y-coordinate, in meters")
ax.set_xlabel("x-coordinate, in meters")
styles.heading(ax, letter="A", heading="Map view")
styles.remove_edge_ticks(ax)
ax = fig.add_subplot(gs[0:5, 6:])
mm = flopy.plot.PlotCrossSection(model=gwf, ax=ax, line={"row": 8})
mm.plot_grid(lw=0.5, color="0.5")
# items for legend
mm.ax.plot(
-1000,
-1000,
"s",
ms=5,
color="green",
mec="black",
mew=0.5,
label="Recharge",
)
mm.ax.plot(
-1000,
-1000,
"s",
ms=5,
color="red",
mec="black",
mew=0.5,
label="Well",
)
mm.ax.plot(
-1000,
-1000,
"s",
ms=5,
color="blue",
mec="black",
mew=0.5,
label="Constant head",
)
mm.ax.plot(
-1000,
-1000,
"s",
ms=5,
color="0.75",
mec="black",
mew=0.5,
label="Inactive",
)
mm.ax.plot(
[-1000, -1001],
[-1000, -1000],
color="orange",
lw=1.5,
label="Cross-section line",
)
# aquifer coloring
ax.fill_between([0, dx.sum()], y1=150, y2=-100, color="cyan", alpha=0.5)
ax.fill_between([0, dx.sum()], y1=-100, y2=-150, color="#D2B48C", alpha=0.5)
ax.fill_between([0, dx.sum()], y1=-150, y2=-350, color="#00BFFF", alpha=0.5)
# well coloring
ax.fill_between(
[dx.cumsum()[8], dx.cumsum()[9]], y1=50, y2=-100, color="red", lw=0
)
# labels
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=300,
y=-97,
text="Upper aquifer",
va="bottom",
ha="left",
fontsize=9,
)
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=300,
y=-147,
text="Confining unit",
va="bottom",
ha="left",
fontsize=9,
)
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=300,
y=-347,
text="Lower aquifer",
va="bottom",
ha="left",
fontsize=9,
)
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=29850,
y=53,
text="Layer 1",
va="bottom",
ha="right",
fontsize=9,
)
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=29850,
y=-97,
text="Layer 2",
va="bottom",
ha="right",
fontsize=9,
)
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=29850,
y=-147,
text="Layer 3",
va="bottom",
ha="right",
fontsize=9,
)
styles.add_text(
ax=ax,
transform=False,
bold=False,
italic=False,
x=29850,
y=-347,
text="Layer 4",
va="bottom",
ha="right",
fontsize=9,
)
ax.plot(
hxloc,
hsxs0,
lw=0.75,
color="black",
ls="dotted",
label="Steady-state\nwater level",
)
ax.plot(
hxloc,
hsxs1,
lw=0.75,
color="black",
ls="dashed",
label="Water-level\nafter period 2",
)
ax.set_ylabel("Elevation, in meters")
ax.set_xlabel("x-coordinate along model row 9, in meters")
styles.graph_legend(
mm.ax,
ncol=2,
bbox_to_anchor=(0.7, -0.6),
borderaxespad=0,
frameon=False,
loc="lower center",
)
styles.heading(ax, letter="B", heading="Cross-section view")
styles.remove_edge_ticks(ax)
if plot_show:
plt.show()
if plot_save:
fpth = figs_path / f"{sim_name}-grid.png"
if not silent:
print(f"saving...'{fpth}'")
fig.savefig(fpth)
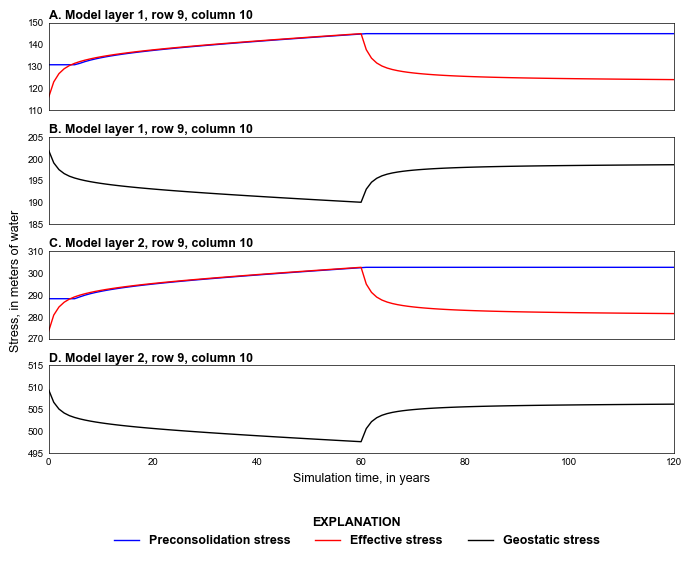
def plot_stresses(sim, silent=True):
with styles.USGSPlot() as fs:
name = sim.name
cd = get_csub_observations(sim)
tmax = cd["totim"][-1]
fig, axes = plt.subplots(
ncols=1,
nrows=4,
figsize=figure_size,
sharex=True,
constrained_layout=True,
)
idx = 0
ax = axes[idx]
ax.set_xlim(0, tmax)
ax.set_ylim(110, 150)
ax.plot(
cd["totim"],
cd["PC1"],
color="blue",
lw=1,
label="Preconsolidation stress",
)
ax.plot(cd["totim"], cd["ES1"], color="red", lw=1, label="Effective stress")
styles.heading(ax, letter="A", heading="Model layer 1, row 9, column 10")
styles.remove_edge_ticks(ax)
idx += 1
ax = axes[idx]
ax.set_ylim(185, 205)
ax.plot(cd["totim"], cd["GS1"], color="black", lw=1)
styles.heading(ax, letter="B", heading="Model layer 1, row 9, column 10")
styles.remove_edge_ticks(ax)
idx += 1
ax = axes[idx]
ax.set_ylim(270, 310)
ax.plot(cd["totim"], cd["PC2"], color="blue", lw=1)
ax.plot(cd["totim"], cd["ES2"], color="red", lw=1)
styles.heading(ax, letter="C", heading="Model layer 2, row 9, column 10")
styles.remove_edge_ticks(ax)
idx += 1
ax = axes[idx]
ax.set_ylim(495, 515)
ax.plot(
[-100, -50],
[-100, -100],
color="blue",
lw=1,
label="Preconsolidation stress",
)
ax.plot([-100, -50], [-100, -100], color="red", lw=1, label="Effective stress")
ax.plot(cd["totim"], cd["GS2"], color="black", lw=1, label="Geostatic stress")
styles.graph_legend(
ax,
ncol=3,
bbox_to_anchor=(0.9, -0.6),
)
styles.heading(ax, letter="D", heading="Model layer 2, row 9, column 10")
styles.remove_edge_ticks(ax)
ax.set_xlabel("Simulation time, in years")
ax.set_ylabel(" ")
ax = fig.add_subplot(111, frame_on=False, xticks=[], yticks=[])
ax.set_ylabel("Stress, in meters of water")
if plot_show:
plt.show()
if plot_save:
fpth = figs_path / f"{name}-01.png"
if not silent:
print(f"saving...'{fpth}'")
fig.savefig(fpth)
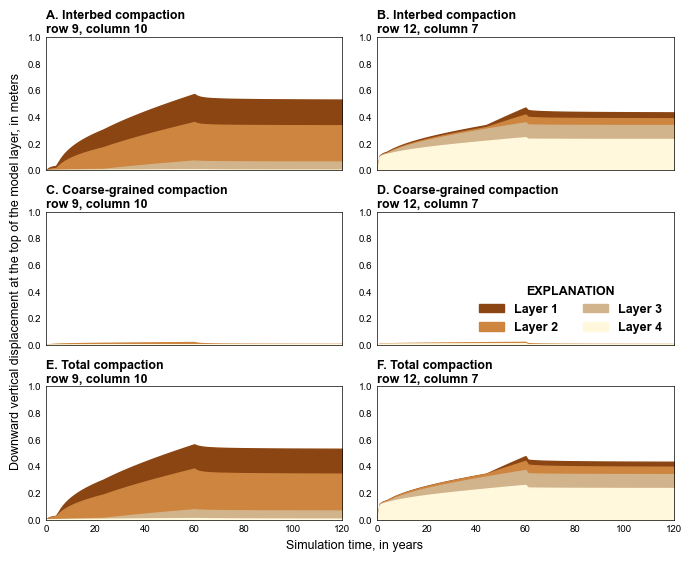
def plot_compaction(sim, silent=True):
with styles.USGSPlot():
name = sim.name
fig, axes = plt.subplots(
ncols=2,
nrows=3,
figsize=figure_size,
sharex=True,
constrained_layout=True,
)
axes = axes.flatten()
idx = 0
ax = axes[idx]
ax.set_xlim(0, 120)
ax.set_ylim(0, 1)
plot_compaction_values(ax, sim, tagbase=plot_tags[idx])
ht = f"Interbed compaction\n{compaction_heading[0]}"
styles.heading(ax, letter="A", heading=ht)
styles.remove_edge_ticks(ax)
idx += 1
ax = axes[idx]
ax.set_ylim(0, 1)
plot_compaction_values(ax, sim, tagbase=plot_tags[idx])
ht = f"Interbed compaction\n{compaction_heading[1]}"
styles.heading(ax, letter="B", heading=ht)
styles.remove_edge_ticks(ax)
idx += 1
ax = axes[idx]
ax.set_ylim(0, 1)
plot_compaction_values(ax, sim, tagbase=plot_tags[idx])
ht = f"Coarse-grained compaction\n{compaction_heading[0]}"
styles.heading(ax, letter="C", heading=ht)
styles.remove_edge_ticks(ax)
idx += 1
ax = axes[idx]
ax.set_ylim(0, 1)
plot_compaction_values(ax, sim, tagbase=plot_tags[idx])
ht = f"Coarse-grained compaction\n{compaction_heading[1]}"
styles.heading(ax, letter="D", heading=ht)
styles.remove_edge_ticks(ax)
styles.graph_legend(ax, ncol=2, loc="lower right")
idx += 1
ax = axes[idx]
ax.set_ylim(0, 1)
plot_compaction_values(ax, sim, tagbase=plot_tags[idx])
ht = f"Total compaction\n{compaction_heading[0]}"
styles.heading(ax, letter="E", heading=ht)
styles.remove_edge_ticks(ax)
ax.set_ylabel(" ")
ax.set_xlabel(" ")
idx += 1
ax = axes.flat[idx]
ax.set_ylim(0, 1)
plot_compaction_values(ax, sim, tagbase=plot_tags[idx])
ht = f"Total compaction\n{compaction_heading[1]}"
styles.heading(ax, letter="F", heading=ht)
styles.remove_edge_ticks(ax)
ax = fig.add_subplot(111, frame_on=False, xticks=[], yticks=[])
ax.set_ylabel(
"Downward vertical displacement at the top of the model layer, in meters"
)
ax.set_xlabel("Simulation time, in years")
if plot_show:
plt.show()
if plot_save:
fpth = figs_path / f"{name}-02.png"
if not silent:
print(f"saving...'{fpth}'")
fig.savefig(fpth)
def plot_results(sim, silent=True):
plot_grid(sim, silent=silent)
plot_stresses(sim, silent=silent)
plot_compaction(sim, silent=silent)
Running the example
Define and invoke a function to run the example scenario, then plot results.
[5]:
def scenario(silent=True):
sim = build_models()
if write:
write_models(sim, silent=silent)
if run:
run_models(sim, silent=silent)
if plot:
plot_results(sim, silent=silent)
scenario()
<flopy.mf6.data.mfstructure.MFDataItemStructure object at 0x7f3601be5100>
run_models took 2455.42 ms
/opt/hostedtoolcache/Python/3.9.19/x64/lib/python3.9/site-packages/IPython/core/pylabtools.py:152: UserWarning: constrained_layout not applied because axes sizes collapsed to zero. Try making figure larger or axes decorations smaller.
fig.canvas.print_figure(bytes_io, **kw)
/tmp/ipykernel_5249/3327239510.py:322: UserWarning: constrained_layout not applied because axes sizes collapsed to zero. Try making figure larger or axes decorations smaller.
fig.savefig(fpth)